BIOP-desktop
The documentation of the BIOP-desktop
| Demo | Installation | Build | Run | FAQ |
Acknowledgements
This whole project was possible thanks to the previous work of the renku team (renku-desktop) and the support of the RCP and SV IT teams, EPFL.
Introduction
The BIOP-desktop is a Versioned Computer with software pre-installed and pre-configured so you can focus on your analysis (and not on the installation).
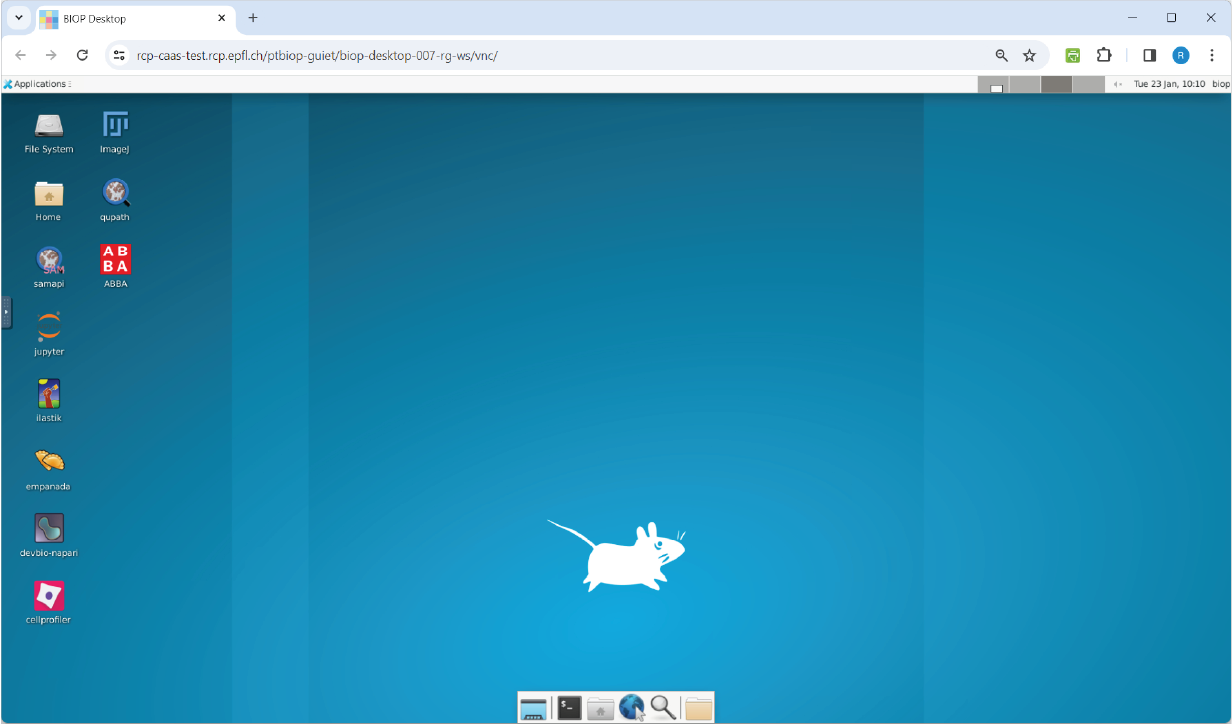
The BIOP-desktop is a Docker image that you can simply pull and run (and optionnaly build yourself)
 ( enlarge image )
( enlarge image )
The BIOP-desktop is :
| Developper Point of View | IT Point of View | Users Point of View |
|---|---|---|
a Docker image that you can build using multi-stage |
a Docker image that you can simply pull and run |
a Computer with “everything” installed and running! |
What is a Versioned Computer?
By Versioned Computer, we mean a computer in which every software is set, making it possible to reproduce the same environment at any time.
Making use of the Docker images, we can create a Versioned Computer that is reproducible, shareable, and open.
We make use of multi-stage build to ease building and versionning. Indeed we can build a new image, changing a single component and keeping the rest of the image unchanged, if needed.
What is a Versioned Computer for?
A Versioned Computer is useful for:
- Focusing on your analysis: you don’t have to worry about installing and configuring software.
- Reproducibility: you can reproduce your analysis at any time, even years later.
- Collaboration: you can share your analysis with your collaborators, and they can reproduce it.
- Teaching: you can share your analysis with your students, and they can reproduce it.
- Publication: you can share your analysis with the reviewers, and they can reproduce it.
- Open Science: you can share your analysis with the world, and they can reproduce it.
Why did we create the BIOP-desktop?
We created the BIOP-desktop to help you focus on your analysis, and not on the installation and configuration of software. BUT we created it also because we need it ourself!
As a core facility, we give courses on image processing and image analysis and practical training on software we create workflows and we wanted a solution that can work locally (on our workstations 12-32 cores, 128-512GB RAM, 12-24GB GPU) and remotely (cluster with 80GB-GPU).
Mainting the software on all our workstation is a burden and we wanted a solution that can ease this task.
BIOP-desktop installed software
| Software | Description | Notes |
|---|---|---|
| ABBA | Aligning Big Brains & Atlases | including : elastix & transformix, DeepSlice, … |
| cellprofiler | Open-source software designed to enable biologists to quantitatively measure phenotypes from thousands of images automatically. | |
| cellpose | An anatomical segmentation algorithm, to detect nucleus, cells, and much more ! | and omnipose too! |
| devbio-napari | The Fijiest way to do image analysis in Python | |
| EMPanADA | DNN to segment mitochondria in EM images | |
| Fiji | A “batteries-included” ImageJ | update sites : clij, stardist, ptbiop , ilastik … |
| ilastik (GPU) | Interactive learning and segmentation toolkit | GPU version |
| inkscape | Vectoriel drawing (and more!) | with inkscape-imagej-panel plugins |
| QuPath | Digital Pathology | with some extensions : cellpose, SAM , Warpy , StarDist… |
| StarDist | Object Detection with Star-convex Shapes |